-
Biosketch
Education
Ph.D.
Institute of Fundamental Sciences, Massey University, New Zealand. Submitted Nov 2009 and Defended Aug - 2010
PhD under Prof. Bill Williams, IFS, Massey University, New Zealand.M.Sc- Physical Chemistry
School of Chemical Sciences, Mahathma Gandhi University, Kerala, India. Dec - 2004
B.Sc- Chemistry
Sir Syed College, Kannur University, Kerala, India. May - 2002Postdoctoral Research
Research Associate ( Dec - 2009 -- May - 2013 )
Lehrstuhl fuer Theoretische Chemie, Ruhr-Universitaet Bochum, Germany.
Research associate with Professor Dr. Dominik Marx. Work was mainly focused on calculating and simulating the mechanochemistry of ring opening reactions and mechanochemical bond breaking mechanisms of protein disulfides. Expertize in QM/MM, Metadynamics and Advanced sampling techniques.Assistant Professor (Adhoc)
National Institute of Technology, Calicut (Feb 2014 to May 2014)
National Institute of Technology, Surathkal (June 2014 to May 2016)Assistant Professor
Indian Institute of Technology, Palakkad (June 2016 - May 2023)
Associate Professor
Indian Institute of Technology, Palakkad (June 2023 onwards)
-
Research
a) Disulfide Bond Cleavage and reaction mechanisms in Proteins under Mechanochemical and Non-Mechanochemical scenario: :

Disulfide bonds play a crucial role in the smooth functioning of protein machinery. Among the proteins, the disulfides are classified into two distinct sets - "Structural" and "Functional" disulfides. The structural disulfides are believed to be added in different protein systems during the evolution to enhance the stability of proteins owing to its strong covalent nature. On the other hand, the comparatively newly defined class of disulfides known as the functional ones are those which are capable of regulating the chemico-biological activity of the protein by themselves undergoing frequent reduction and oxidation. These disulfide bonds are enriched by the very chemistry of sulfur bonds, which can easily undergo redox reactions at mild, physiological conditions. Thus, studying the cleavage reactions and the reformation of the disulfides in proteins becomes extremely crucial as it could be deterministic in defining distinct biological processes.
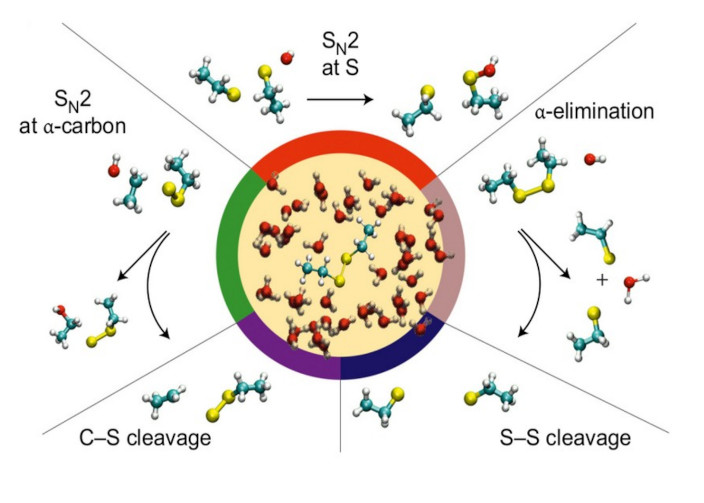
Activation free energies as a function of external force (panel a) for nucleophilic cleavage of diethyl disulfide in bulk water by OH− nucleophile, using isotensional ab initio metadynamics sampling (red) and thermodynamic integration (black) at T = 300 K. Free energy surfaces in the absence of force (panel b) and at a force of 1.5 nN (panel c). CV1 is the difference between the S–S distance (panel d) and the distance of the attacked sulfur, S⋆, with respect to the attacking hydroxyl oxygen, thus (negative and positive) the values corresponding to reactant and product states respectively. CV2 is the S⋆-S-C-C dihedral angle on the side of the reaction centre (where the dotted horizontal lines (panel c) mark 180◦ and thus the so-called closed conformation of diethyl disulfide). The arrows and crosses highlight schematically the energetically preferred and restricted pathways, respectively, according to the topology of the underlying free energy landscape. Red circles (in panel a) mark the points for which the free energy surfaces are shown in panels b and c. Reprinted from P. Dopieralski, J Ribas-Arino, P.Anjukandi et al, Nat. Chem., 2013. Copyright 2013 NPG.
A. G. Nair, D. S. Parumalla, P. Anjukandi*, Submitted, 2021.
P. Dopieralski, J Ribas-Arino, P. Anjukandi et al, Nat. Chem., 2017.
P. Dopieralski, J Ribas-Arino, P. Anjukandi et al, Angew. Chem. Int. Ed. , 2016.
P.Anjukandi*, P. Dopieralski, J Ribas-Arino et al, PLoS One, 2014.
P. Dopieralski, J Ribas-Arino, P. Anjukandi et al, Nat. Chem., 2013.c) Folding Mechanisms in Proteins:
It is great to see how nature organises the packaging of its sub units to make meaningful and yet fully functional working units out of them. Proteins, thought to be one of the fundamental building blocks of life is one such example of its kind. One of the interesting aspect of the proteins are the formation of different structural types while packing themselves.
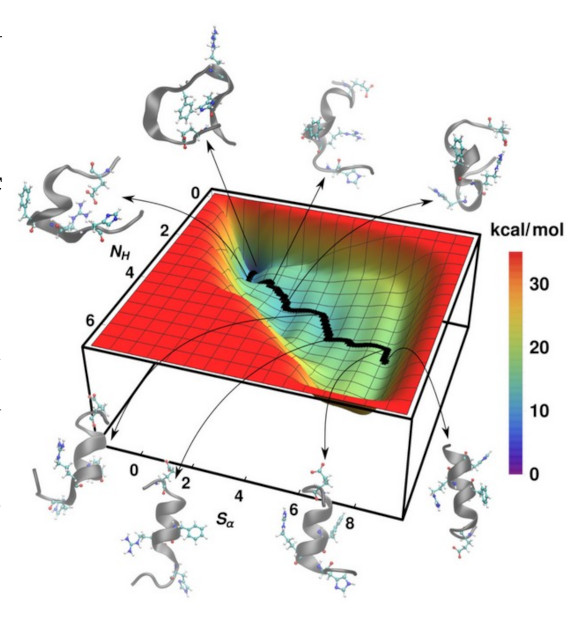
Herein, we are interested basically in understanding the folding, unfolding mechanisms and kinetics in the α-helices and β-hairpin loops, which forms a part of the secondary motifs in proteins.
D. S. Parumalla, G. Govind, P. Anjukandi*, ChemistrySelect, 2020.
G. Govind, E. C. Nayana, P. Anjukandi*, Journal of Biomolecular Structure & Dynamics, 2021.c) Mechanochemical Ring Opening Reactions:
The role played by long side chains (like polyethylene oligomers) in transducing external tensile forces on to different mechanophores plays a great role in defining the chemistry of these mechanophores. Herein, we demonstrate that the oligomer chains do indeed exert a notable influence on the force dependence of the activation energies of ring openings in benzocyclobutene moeities, with both conrotatory and disrotatory ring-opening processes being effected considerably. This could be instrumental in tuning the properties of mechanoresponsive materials not only by changing the properties of the mechanophore itself, but also by tailoring the force-transducing chain molecules attached to it. It is found that these chains even have a profound impact on the topology of the force-transformed potential energy surface in the vicinity of conrotatory transition states.

P. Dopieralski, P. Anjukandi, M. Rauckert et al, J. Mat. Chem. , 2011.
d) Mechanochemistry of Pectins:
Calculating and simulating the mechanical behavior of single biopolymer (Pectin) chains. DFT calculations on monomers, diamers and trimers of pectin to look at their conformations during force induced stretching and characterization of the biopolymer. Brownian Dynamics Simulations on larger chains to look at stretching and the collapse of the chain in different solvent and chain stiffness. Steered Molecular Dynamics (SMD), looking at the stretching of a ten member polymer chain and its force-extension behavior.
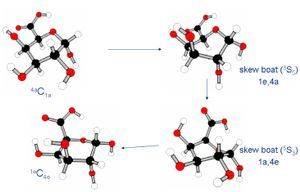
P.Anjukandi G G Pereira and M A K Williams, J. Theo. Biol, 2010.
M.A.K. Williams, A.T. Marshall, P. Anjukandi and R. G. Haverkamp, Phys. Rev. E, 2007. -
Teaching
Joined IIT Palakkad from July 2016 onwards as Assistant Professor and since then, I have been offering Chemistry Theory Courses and Chemistry Lab Courses for both UG and PG students.
• CY1011 (CY1010 previously): This course mainly deals with Thermodynamics, Kinetics and Organic Chemistry and this is UG level course for the B. Tech students.
• CY1021 (CY1020 previously): This is a UG level core course for the B. Techs which mainly focuses on Theoretical and Inorganic Chemistry.
• CY1140 (CY1030 previously): This is an undergraduate lab course for the B. Tech Students.
• CY5005: This is a PG level core course and this course mainly incorporates Symmetry, Group Theory and Spectroscopy.
• CY5103: This is an advanced level lab course for the PG students and in here, we have introduced introduction and hands on problems in Computational Chemistry for the UG students.
• CY3601: This elective course mainly associates with Electrochemistry and Corrosion and is offered for the 5th and 7th semester B. Tech students.
• CY5609: This is an advanced level Quantum Chemistry course which incorporates both the theory as well as the lab and is offered mainly oriented towards the PG community.
Apart from these, I have been actively offering the internships and project (internal/external) for the UG and PG students.
-
Research Group
Present Group Members
Research Associate:
Enthusiastic people interested in applying for N-PDF and other national fellowships for research positions may kindly contact.
Ph.D. Students:
Date of Joining : June 2017
Research Work: Towards the Mechanism of Different Disulfide Bond Cleavages in Proteins.
Academic Background:
- Gate, 2016.
- M.Sc, Amrita School of Arts and Science, Kollam, 2012.
- B.Sc, S D College, Alappuzha, 2010.
Date of Joining : June 2019
Research Work:
Academic Background:
Gate, 2019.
M.Sc, Payyannur College, Kannur, 2015.
B.Sc, Sir Syed College, Kannur, 2013.
Research Assistant:
Will be updated soon.
Past Group Members
Research Associate:
1) Dr. D Sravankumar Parumalla, currently a PDF at IPC, IISc, Bangalore.
Research Assistant:1) Mr. Gokul Govind, Presently a Ph.D student at Institute of Physics, Johannes Gutenberg Universitat Mainz, Germany.
M.Sc Project Students from IIT Palakkad:
1) Mr. Anurag Pandey.
Master Project (MSc 2 Year), 2021.
Project Title: Computational study of interaction of the C4 domain of VWF Protein with Silver Nanoparticles
2) Ms. Ishmita Kaur.
Master Project (MSc 2 Year), 2021.
Project Title: Studies of single G-T mismatch in A-T rich DNA hairpins using AMBER03 force-field methods: comparison with G-C match base pair
3) Ms. Rimisha A. C.
Master Project (MSc 2 Year), 2021.
Project Title: Study of different Nickel complexes using density functional theory.M.Sc Project Students and Internship Students form other Institutes:
1) Ms. Amirtha N. R., Department of Chemistry, NSS College, Nemmara, Calicut University.
Master Project (MSc 2 Year), Summer 2017.
Project Title: Optical Properties of π-Conjugated Materials are not Merely their Molecular Properties
2) Ms. Aswathy P. S., Department of Chemistry, NSS College, Nemmara, Calicut University.
Master Project (MSc 2 Year), Summer 2017.
Project Title: Conformations are Deterministic in Defining the Underlying Potential Energy Surface of Mechanochemical Reaction3) Ms. Athira K, Department of Physics, MES College, Marampally, M G University.
Master Project (MSc 2 Year), Summer 2018.
Project Title: Screening π–Conjugated Organic Materials for Thermally Activated Delayed Fluorescence4) Ms. Sona C P, Department of Physics, MES College, Marampally, M G University.
Master Project (MSc 2 Year), Summer 2018.
Project Title: Screening π–Conjugated Organic Materials for Thermally Activated Delayed Fluorescence5) Mr. Antony K Joseph, Department of Chemistry, Victoria College, Palakkad, Calicut University.
Master Project (MSc 2 Year), Summer 2018.
Project Title:6) Ms. Celin Rooth, Department of Chemistry, St. Alberts College, Ernakulam, M G University. (Presently a Ph.D student at Chemistry Department, IIT Madras)
Master Project (MSc 2 Year), Summer 2018.
Project Title: Towards the Conductivity of Ferrocene Based Nanowires.7) Mr. Gokul Govind, Department of Chemistry, Central University, Thiruvarur, Tamil Nadu.
Summer Intern (Integrated M.Sc 3 Year), Summer 2018.
Project Title: Urea Depended Denaturation of ß-Hairpin of B1 domain of Protein G - A Molecular Dynamics Study8) Mr. Rinto Thomas, Department of Computational Sciences, Central University of Punjab.
Summer Intern (M.Sc 1 Year), Summer 2019.
Project Title: Self Aggregation of Hydrogen Cyanide: A molecular Dynamics study9) Ms. Archana K, PSGR Krishnammal College for Women, Coimbatore, Tamil Nadu.
Summer Intern (M.Sc 1 year) from Indian Academy of Sciences, Summer 2019.
Project Title: Electrochem- a Graphical User Interface(GUI) using MATLAB10) Ms. Archana K, PSGR Krishnammal College for Women, Coimbatore, Tamil Nadu.
Master Project (MSc 2 Year), Summer 2020.
Project Title: MD Simulation Study on DSBC and DSBC-nDSBD Complex Proteins.11) Mr. Gokul Govind, Department of Chemistry, Central University, Thiruvarur, Tamil Nadu. (Presently a Ph.D student at Institute of Physics, Johannes Gutenberg Universitat Mainz, Germany.)
Master Project (Integrated M.Sc 5 Year), Summer 2020 (Jan 2020 - May 2020).
Project Title: Insights into the Folding–Unfolding Dynamics of pH-Assisted Structures of Ribonuclease S-Peptide. -
Additional Information
TitleHobbyDescription
Photography, Gardening and Cooking.
-
Publications
Lavanya Rao, John D Rodney, Anjalin Joy, Chadva Shivangi Nileshbhai, Anupriya James, Sushmitha S, Fiona Joyline Mascarenhas, N.K. Udayashankar, Padmesh Anjukandi*, Byung Chul Kim*, Badekai Ramachandra Bhat*Chemical Engineering Journal 500 156639 (2024)Aparna G Nair* and Padmesh A*J. Phys. Chem. B 128 (43) 10541-10552 (2024)Shyama Ramakrishnan* and Padmesh Anjukandi*Inorg. Chem. 63 (32) 15186-15196 (2024)Nayana Edavan Chathoth*, Hafila Khairun S, Manya Krishna and Padmesh Anjukandi*J. Mater. Chem. B 12 3908-3916 (2024)Aparna G Nair, Arathi Das, Nayana Edavan Chathoth, Manash Pratim Sarmah and Padmesh Anjukandi*ChemPhysChem Just Accepted (2023)Nayana E C, Aparna G Nair, and Anjukandi Padmesh*PCCP (2023)Anjitha Thadathil, Nayana Edavan Chathoth, Yahya A. Ismail, Padmesh Anjukandi*, and Pradeepan Periyat*J. Phys. Chem. C 126 (40) 16965–16982 (2022)Aparna G Nair, D. Sravanakumar Perumalla, Padmesh Anjukandi*ChemPhysChem Accepted (2022)Aparna G Nair, D. Sravanakumar Perumalla and Padmesh AnjukandiPhysical Chemistry Chemical Physics 24 7691 - 7699 (2022)Gokul Govind, Nayana E C, Padmesh AJournal of Biomolecular Structure & Dynamics 1-7 (2021)D. Sravanakumar Perumalla, Gokul Govind and Padmesh AnjukandiChemistrySelect 5 5748-5755 (2020)Przemyslaw Dopieralski, Jordi Ribas-Arino, Padmesh Anjukandi, Martin Krupicka, and Dominik MarxNature Chemistry 9 164-170 (2017)Przemyslaw Dopieralski, Jordi Ribas-Arino, Padmesh Anjukandi, Martin Krupicka, and Dominik MarxAngew. Chem. Int. Ed. 55 1304-1308 (2016)P. Anjukandi*, P. Dopieralski, J. Ribas-Arino and D. Marx;PLoS One 9 (10) e108812 (2014)P. Dopieralski, J. Ribas-Arino, P. Anjukandi, M. Krupicka, J. Kiss, and D. MarxNature Chemistry 5 685-691 (2013)P. Dopieralski, Padmesh Anjukandi, Jordi Ribas-Arino and Dominik MarxinSiDE 10 48-52 (2012)Abdenor Fellah, Padmesh Anjukandi, Yacine Hemar, Don Otter and Martin A.K. WilliamsCarbohydrate Polymers 86 (1) 105-111 (2011)Przemyslaw Dopieralski, Padmesh Anjukandi, Matthias Ruckert, Motoyuki Shiga, Jordi Ribas-Arino and Dominik MarxJournal of Materials Chemistry 21 8309-8316 (2011)Renji R. Reghu, Hari Krishna Bisoyi, Juozas V. Grazuleviciu, Padmesh Anjukandi, Valentas Gaidelis and Vygintas JankauskasJournal of Materials Chemistry 21 7811-7819 (2011)Padmesh Anjukandi, Gerald G. Pereira and Martin A.K. WilliamsJournal of Theoretical Biology 265 (3) 245-249 (2010)Abdenor Fellah, Padmesh Anjukandi, Mark Waterland and Martin A.K. WilliamsCarbohydrate Polymers 78 (4) 847-853 (2009)M.A.K. Williams, A.T. Marshall, P. Anjukandi and R. G. HaverkampPhysical Review E 76 021927 (2007)
INDIAN INSTITUTE OF TECHNOLOGY PALAKKAD
Kanjikode | Palakkad | Kerala - 678623
Kanjikode | Palakkad | Kerala - 678623
0491 209 2001 (Office) |
info@iitpkd.ac.in
Copyright ©2023 Indian Institute of Technology Palakkad. All Rights Reserved.


